Brain blood flow can be measured in many ways, but in this post I will focus on transcranial Doppler ultrasound (TCD). This method is used to measure the speed and direction of blood flow through a single blood vessel in your head.
TCD is a great measure for brain blood flow: the method can measure blood flow beat-by-beat (we tend to say that it has excellent temporal resolution – it collects a lot of data points per unit time). It is also non-invasive, meaning it causes no harm to the body (although a lower power should be used for some probe placements). It is very good for measure changes in blood flow, for example due to certain stimuli, and can be used to measure how well blood flow responds to such stimuli.
So what is this signal? What happens when we use ultrasound is that an ultrasonic beam is sent from the probe and into the blood vessel, where it is reflected back from the moving red blood cells. The returned signal is slightly different from the first signal. If the blood cells are moving towards the probe, the returned signal has a higher frequency. If it is moving away from the probe, the returned signal has a lower frequency. How much of a faster or lower signal depends on how fast the blood cells move. We call this change in wavelength the Doppler shift or the Doppler effect, and it has the symbol f. It is this Doppler shift that is being used to calculate blood flow velocity.
Most of us have seen the Doppler shift in action. A common example is if an ambulance or fire engine passes us in the street with its sirens on. When it comes towards us, the source of the sound (the vehicle) is moving in the same direction as the sound of the siren, and the sound waves are therefore compacted (have a shorter wavelength), resulting in a higher frequency sound (a higher pitch). When it has passed us and moves away, the vehicle moves in the opposite direction to the sound of the siren (relative to us) and so the sound waves are stretched (have a longer wavelength), we get a lower frequency sound (a lower pitch).
(Not clear? Here is a Vimeo video illustrating the Doppler effect and the example above)
Ok, which blood vessel is best. There are plenty of blood vessels in the brain, but the middle cerebral artery (MCA) is great as it is a major artery and easy to locate. The MCA emerges from the carotid artery, which is the main artery going to the head. We have one carotid artery on each side of the neck (which you can feel when you take your pulse). At the top of the neck, the carotid artery divides in an internal and an external branch. The internal branch then divides into the MCA and the anterior cerebral artery (ACA). The MCA receives around 70% of the internal carotid artery blood flow, and we therefore often assume that blood flow through the MCA is representative of the total blood flow to one half of the brain.
How to measure the flow. When we image blood flow through this vessel using TCD, we put the ultrasound probe to a person’s temple, and choose an ultrasound beam depth at around 45-60 mm. The best depth depends on the anatomy of the person – there is a bit of variation in the population, so you may have to try different depths to get a good signal. The temple is a good place to measure blood flow, as it is where the internal cerebral artery splits into the ACA (blood flowing away from the probe) and the MCA (blood flowing towards the probe). This gives a very distinct waveform (see below) and means we can be reasonably sure we have placed the probe and the ultrasound beam correctly.
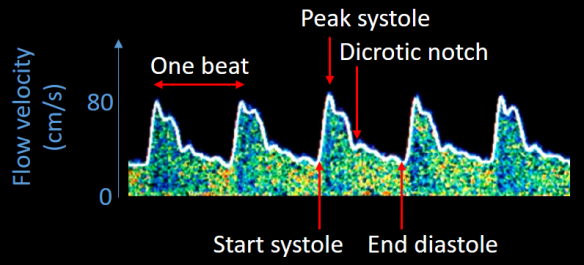
Typical TCD trace from the middle cerebral artery.
It should be mentioned that when measuring blood flow through other vessels, we may choose a different placement of the probe. This can include putting the probe over the (closed) eye (transorbital), at the base of the neck (suboccipital) or on the neck below the ear (submandibular). We call these placements ‘windows’ (i.e. the transtemporal window is the placement of the probe on the temple). From the transtemporal window, we can see flow through the MCA, ACA, the terminal internal cerebral artery (ICA) and the posterior cerebral artery (PCA).
What is the physiology behind the signal? The waveform shows changes in blood flow due to systolic and diastolic phases of the heart. Referring to the image above, each one of the ‘waves’ is a heartbeat. The start of the systole, peak systole, dicrotic notch and the end of the diastole are marked on the figure. Systole is when the heart contracts and pumps the blood out, and diastole is when the heart relaxes and refills with blood. The dicrotic notch is a short-term change in aortic pressure that stems from the closure of the aortic valve. This valve is between the left ventricle of the heart and the aorta, and blood passes this valve as it enters the body circulation.
So it starts with the beginning of the systole (when the heart starts to contract). The increased pressure in the contracting heart pushes blood out of the left ventricle and into the circulation through the aorta. Some of this blood goes to the brain and is what we measure with TCD. Blood flow increases through the blood vessels as it is being pumped out of the heart. At peak systole, the blood flow for that particular heartbeat is at its maximum. Then, as the heart begins to relax, the pressure in the ventricle drops and it begins to refill with blood (coming from the lungs). The aortic valve closes and stops blood from flowing the wrong way back into the heart. This closure of the valve can be seen as a small change in blood flow (the dicrotic notch). We reach the lowest flow just as the heart is filled up and ready to contract again. TCD gives us a measure of the blood flow at all these stages of the cardiac cycle.
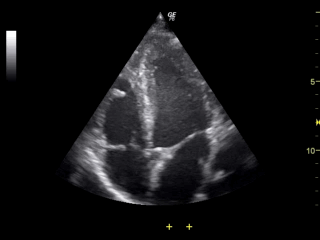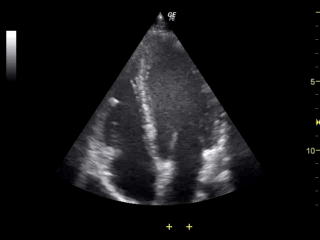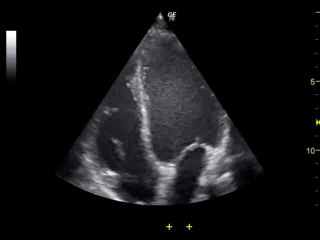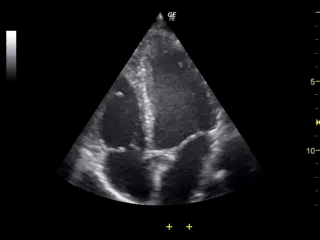
Ultrasound of the heart. All four heart chambers are visible, as are the valves (flapping open and closed) between the atrial and ventrical chambers.
The temporal resolution of TCD is excellent. As can be seen in the gif above, ultrasound lets us see things in more or less real time (in this case a beating heart), and this makes it a useful technique for measuring rapid changes in brain blood flow.
I will be posting a short tutorial on how to analyse TCD data soon, and if anyone has any questions on the method before then, I would be happy to address them in the comments.